Estimation of ECM Signatures in Fiber Probe Images
Developing a novel deep learning framework to automate Extracellular Matrix (ECM) quantification from noisy fiber probe imagery.
This project is currently being developed as the Final Year Research Project at the Department of Electronic and Telecommunication Engineering, University of Moratuwa.
Project Idea
Traditional histopathology relies on tissue excision (biopsy) and staining, limiting it to ex-vivo analysis. Our project leverages domain adaptation to quantify collagen fiber organization directly from label-free fiber probe images, enabling in-vivo assessment without biopsies.
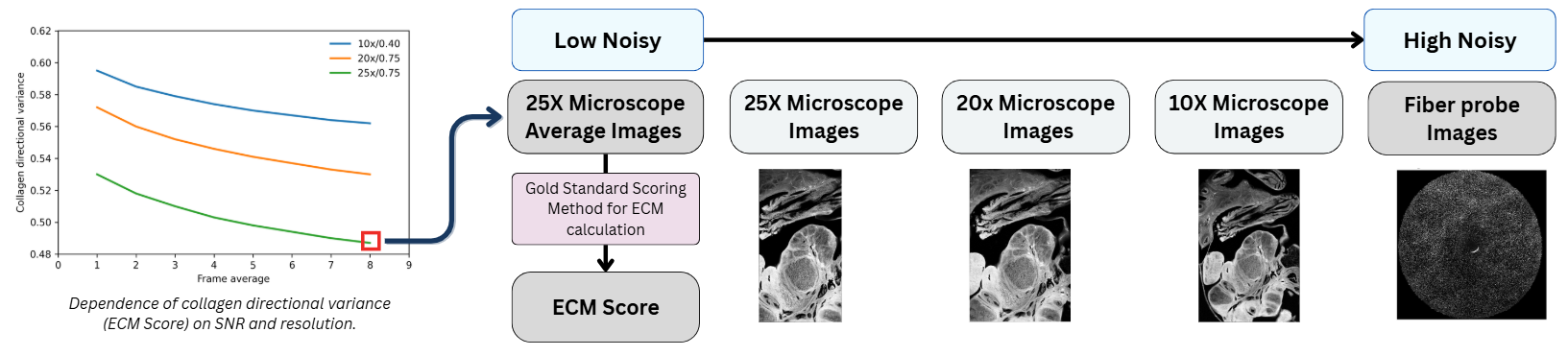
Problem Significance
The structural organization of the Extracellular Matrix (ECM), particularly collagen fibers, is a strong indicator of cancer malignancy. Current intra-operative tools like frozen section analysis can delay procedures and are subject to sampling errors. There is a critical need for a tool that provides immediate feedback on tissue status during surgery.
Problems with Existing Solutions
Standard table-top SHG microscopes are bulky and restricted to ex-vivo samples. While fiber-optic probes offer portability, they suffer from degraded image quality, including low resolution, low Signal-to-Noise Ratio (SNR), and artifacts. Standard computer vision models fail to generalize due to the significant domain shift between high-quality microscope images and noisy probe inputs.
Our Solution
Our solution implements a comprehensive five-stage deep learning pipeline designed to transform raw optical data into clinically relevant ECM predictions:
- Stage 0: Preprocessing & Ground Truth Generation: Establishes the regression targets by co-registering microscopic frames and calculating “Gold Standard” Directional Variance (DV) scores from high-resolution imagery.
- Stage 1: Multi-resolution Encoder: Trains a foundation model using Rank-N-Contrast (RnC) Loss to learn a robust, magnification-invariant, and regression-aware embedding space.
- Stage 2: Task Head & Reconstruction: Freezes the encoder to train a lightweight MLP regressor for ECM scoring, while simultaneously training a Decoder to ensure feature preservation through image reconstruction.
- Stage 3: Cross-Modality Registration: Bridges the domain gap between high-quality microscopy and noisy fiber probes using CycleGAN and surrogate modeling to translate domains without paired data.
- Stage 4: Encoder Finetuning: Aligns the fiber probe embeddings with the ground truth microscope representations to enable direct, accurate inference on real-world probe images.
My Role
I led the research and development of Stage 3: Cross-Modality Registration, focusing on the generative AI components required to bridge the domain gap between high-fidelity microscopy and noisy fiber probe imagery.
My Contribution
My key contributions include:
- CycleGAN Implementation: Designed and optimized a CycleGAN architecture for unpaired image-to-image translation, enabling the generation of realistic fiber probe training data from microscope images.
- Surrogate Modeling: Developed a PSF-based blur model to mathematically simulate optical degradation, providing physically grounded initialization for the GAN.
- Architecture Optimization: Integrated LSGAN loss and Spectral Normalization to stabilize training and preserve the specific circular field-of-view texture of the fiber probes.
Architecture
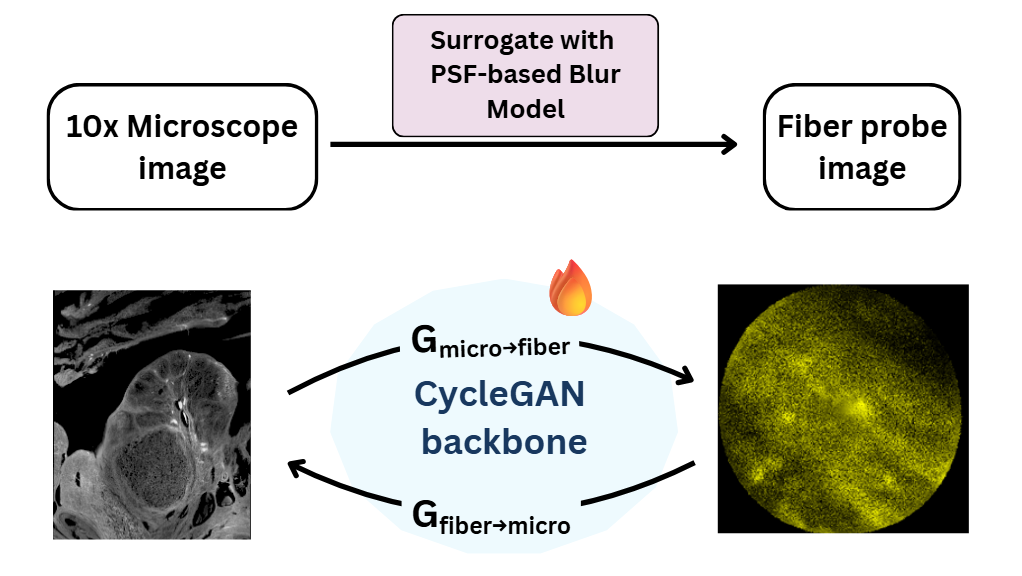
The architecture above illustrates the Cross Modality Registration model I developed. It features a dual-generator setup (\(G_{micro \rightarrow fiber}\) and \(G_{fiber \rightarrow micro}\)) initialized with a surrogate PSF model. This allows us to “hallucinate” realistic probe artifacts onto clean data for training, and conversely, restore clean structures from noisy inputs for analysis.
Impact
This tool validates that clinical-grade ECM quantification does not require microscope-quality images. It enables real-time, intra-procedural guidance for tumor margin assessment, facilitating faster diagnosis and potentially better surgical outcomes.
Supervisors
This research is conducted under the guidance of:
- Dr. Chamira Edussooriya (University of Moratuwa, Sri Lanka)
- Dr. Ranga Rodrigo (University of Moratuwa, Sri Lanka)
- Dr. Dushan Wadduwage (Old Dominion University, USA)
- Dr. Einstein Gnanatheepam (Tufts University, USA)
This project is actively under development as part of my Final Year Research at the University of Moratuwa. Comprehensive technical details, experimental validation, and quantitative performance metrics will be made available upon completion of the upcoming publication.